>Corresponding Author : Thomas M Johnson
>Article Type : Original Research Article
>Volume : 4 | Issue : 1
>Received Date : 20 Jan, 2024
>Accepted Date : 01 Feb, 2024
>Published Date : 06 Feb, 2024
>DOI : https://doi.org/10.54289/JDOE2400103
>Citation : Retrum JK, Washington MA, Boudreaux DM, Lincicum AR, Stancoven BW, et al. (2024) The Influence of Hydroxyapatite, Titanium, and Yttria-Stabilized Zirconia Oxide on Diversity of Bacterial Cultures Grown from Human Dental Biofilm Specimens in an In Vitro Model. J Dent Oral Epidemiol 4(1): doi https:// doi.org/10.54289/JDOE2400103
>Copyright : © 2024 Retrum JK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Original Research Article | Open Access
1Department of Periodontics, Army Postgraduate Dental School, Postgraduate Dental College, Uniformed Services University, Fort Eisenhower, Georgia, USA
2Department of Clinical Investigation, Dwight David Eisenhower Army Medical Center, Fort Eisenhower, Georgia, USA
3Multidrug-Resistant Organism Repository and Surveillance Network, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA
*Corresponding author: Thomas M Johnson, Department of Periodontics, Army Postgraduate Dental School, Postgraduate Dental College, Uniformed Services University, Fort Eisenhower, Georgia, USA
Abstract
Background: Titanium (TI) and yttria stabilized-zirconia oxide (YSZ) are dental materials commonly utilized at the soft-tissue interface surrounding dental implants. The influence of these surfaces on bacterial adhesion and biofilm development may affect clinical performance and patient susceptibility to inflammatory peri-implant disease. The purpose of this study was to evaluate the influence of the substrate material on biofilm diversity.
Methods: Biofilms were cultured on TI, YSZ, and hydroxyapatite (HA) surfaces (control) using plaque specimens obtained from three human donors. Duplicate cultures grew for one, two, three, six or nine days. Biofilm diversity was then analyzed using 16S rRNA sequencing. The Shannon Diversity Index (SDI) was calculated for each experimental group. Microbial profiles were intercompared in a pairwise fashion to establish dissimilarity scores, which were recorded in a distance dissimilarity matrix.
Results: A total of 16 taxa were identified, and relative abundances of the predominant phyla and genera did not appear statistically different across experimental groups. Biofilms grown on HA surfaces exhibited significantly higher alpha diversity compared with those formed on TI or YSZ (p<0.0001), although biofilms cultured on TI and YSG surfaces exhibited comparable diversity. Statistically significant differences in beta diversity associated with substrate (p=0.012) and growth period (p=0.001) were detected.
Conclusions: Under the conditions described, biofilms grown on TI or YSZ appeared significantly less complex than those formed on HA. Transmucosal implant abutment surface characteristics represent one modifiable factor potentially influencing risk of peri-implant disease. Among risk of peri-implant disease. Among multiple considerations in abutment design, biofilm diversity performance may represent a clinically relevant benchmark.
Keywords: Dental Implants; Biofilms; Dental Plaque; Peri-Implantitis; Mucositis; Titanium
Abbreviations: TI: Titanium, YSZ: Yttria Stabilized-Zirconia Oxide, HA: Hydroxyapatite, SDI: Shannon Diversity Index, JE: Junctional Epithelium, CT: Connective Tissue, DGJ: Dentogingival Junction, WRAIR: Walter Reed Army Institute of Research, SDI: Shannon Diversity Index, OTU: Operational Taxonomic Unit, ASV: Amplicon Sequence Variant, PCA: Principal Component Analysis, GC: Guanine-Cytosine, LAD: Leucocyte Adhesion Deficiency, LAP: Localized Aggressive Periodontitis
Introduction
The presence of intact skin and mucosal barriers to protect against invading microbial pathogens ranks among the most fundamental requirements for human health. “Mucosa” refers to an epithelial layer together with the immediately subjacent lamina propria [1]. Such tissues line the respiratory, urogenital, and alimentary tracts. A compromise of the body’s protective epithelial barrier renders the host exquisitely susceptible to infection. The oral cavity presents a unique challenge to the maintenance of an effective mucosal defense. Only in this region of the body is a mucosal barrier interrupted by hard tissue under normal circumstances. In fact, clinicians have considered the dentogingival interface a locus minoris resistentiae (body region more vulnerable than others), highlighting increased potential for microbial ingress and disease progression at this soft-hard tissue boundary [2]. Fortunately, specialized supracrestal attached tissues, comprised of the junctional epithelium (JE) and the connective tissue (CT) attachment, form an effective biologic seal around teeth, supporting health despite discontinuities in the oral mucosa [1,3,4]. During development, the primary epithelial attachment derives from the reduced enamel epithelium, and over time, the JE gradually adopts a stratified squamous appearance [1,4]. Analogous to epithelial-CT interfaces found in tissues throughout the body, the JE attaches via hemidesmosomes and a basal lamina to enamel, cementum, or dentin at the dentogingival junction (DGJ) [1,3-5]. Deep to the JE, multiple gingival fiber groups insert into the root surface as Sharpey’s fibers, forming the CT attachment [1,3]. The terminal ends of these densely packed extrinsic fibers are invested in cementum and mineralized, firmly anchoring the CT to the tooth [4].
Dental implants and associated components, which functionally replace missing teeth, also extend through the oral mucosa and thus carry the same requirement for a biological seal. In some respects, soft tissue interfaces at implant sites and teeth are similar. In both circumstances, hemidesmosomes and basal laminae mediate the epithelial attachment [4]. However, implants are biocompatible inorganic devices installed in host bone following tooth loss or agenesis, whereas natural teeth are products of a complex developmental process that proceeds in concert with establishment of periodontal tissues. Thus, a transmucosal implant abutment lacks the organic connective tissue attachment found at the DGJ [4]. Instead, connective tissue merely approximates the abutment material with collagen fibers predominantly oriented parallel to the surface of the device [4].
Both teeth and implants function within a complex oral environment, and the soft tissue interfaces at both site types are under constant microbial challenge. Periodontitis ranks as the most prevalent noncommunicable chronic inflammatory disease affecting humans[6-8], and peri-implant diseases appear comparably common, meta-analyses having estimated the patient-level prevalences of peri-implant mucositis and peri-implantitis at 43% and 22%, respectively [9]. Periodontal and peri-implant diseases share a common etiology—microorganisms found within dental biofilms—and the pathogenesis of both diseases involves inflammatory destruction of host tissue [10].
The structure of dental plaque and the processes by which dental biofilms develop and mature are well known, but questions regarding the precise etiological basis of periodontitis and peri-implantitis remain [11,12]. Members of the Streptococcus and Actinomyces genera exemplify early gram positive colonizers that adhere to acquired pellicle proteins coating tooth surfaces [13,14]. Fusobacterium species exhibit the unusual ability to engage in pairwise coaggregations with a wide variety of partner strains, including typical early colonizers in dental biofilms [15]. The complex of bacteria strongly associated with clinical signs of periodontal tissue destruction—Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola—is rarely isolated in the absence of species such as Fusobacterium nucleatum, which link this “red complex” with early colonizers [16]. Currently, the prevailing conception of periodontal/peri-implant disease initiation and progression involves a process known as dysbiosis—the tipping of a health-associated microbial community toward a biofilm composition not adequately countered by the host immune response [11,12]. This loss of homeostasis, which leads to tissue destruction, can manifest as a decrease in microbial diversity, an increase in pathogenic species, or a contraction in beneficial microorganisms [12]. An emerging hypothesis posits that periodontitis and peri-implantitis are not caused by one or a few essential bacterial species. Rather, disease results from true polymicrobial activity involving accumulation of specific combinations of genes, and thus functions, within the biofilm [11].
In treated periodontitis patients, ample evidence suggests that mechanical disruption of biofilms through sound oral hygiene practices and a monitored professional maintenance program effectively produces periodontal health and stability for most individuals [17-19]. Moreover, dental plaque removal will reverse or prevent gingivitis in non-periodontitis patients [20]. Effective oral hygiene is equally critical for the maintenance of peri-implant health [21,22]. Factors that impede effective plaque removal have been associated with peri-implant mucositis and peri-implantitis [21-24].
One determinant of oral hygiene effectiveness at a dental implant site is the specific material selected for the transmucosal abutment. The substrate represents a major factor influencing adhesion of bacterial and host cells. The process of bacterial adhesion involves both physicochemical and molecular interactions [25]. Adhesion to abiotic substances typically involves nonspecific interactions, whereas specific ligand-receptor interactions predominate in bacterial adhesion to biological surfaces [15,26-28]. Bacterial adhesion is an early step in biofilm development, and decreasing bacterial adhesion may promote health and stability of peri-implant tissues. The purpose of this in vitro study was to compare the complete microbiomes cultured on three substrates— HA, TI, and YSZ—subjected to human dental biofilms ex vivo.
Materials And Methods
This protocol utilized de-identified dental biofilm and saliva specimens and did not involve contact with patients or patient records. Thus, the Dwight David Eisenhower Army Medical Center Human Research Protections Office determined this research to be exempt from Institutional Review Board review requirements (protocol #20-11855/934337) in accordance with exemption criteria described in 32CFR§219.104(d), Category 4. For this research, use of three de-identified subgingival dental plaque specimens and three matched saliva specimens were requested from the Fort Eisenhower Saliva and Dental Plaque Repository (protocol #RHC-A-20-059). Specimen donors were periodontally healthy nonsmokers who had reported no use of any antibiotic or probiotic agent within 90 days of specimen collection. In each experiment, matched plaque and saliva specimens derived from a single donor.
Substrates
The materials evaluated in this investigation were 9.5-mm diameter discs (2 mm thickness) composed of HA (Clarkson Chromatography Products, South Williamsport, Pennsylvania, USA), 99% pure TI (American Elements, Los Angeles, California, USA, #TI-M-O2-D), and YSZ (American Elements, #ZRO-YSZ-O2R-D). Average surface roughness (Ra) values for all discs were recorded using the Perthometer M2 with PFM Drive Unit (Mahr, Providence, RI, USA). Then, Ra values were modified using an aluminum oxide sandblasting unit to an approximate value of 0.2 µm to minimize roughness-related adhesion variability across the experimental groups.
Bacterial growth
Plaque specimens were gently sonicated in a water bath for 30 seconds to disrupt the biofilm. Sterile HA, TI, and YSZ discs were separated and placed into sterile Petri dishes, then coated for 2 hours at room temperature with 1:10 saliva to allow pellicle formation. Next, the saliva was removed, and 2 mL of a sterile tryptic soy broth supplemented with 5 μg/mL hemin and 1 μg/mL menadione (TSB-h) was gently added to the Petri dishes. Plaque specimens were resuspended by briefly vortexing, and each Petri dish was inoculated with 50 μL of the plaque suspension. The dishes then remained in an anaerobic incubator at 37º C (75%N2/10%CO2/10%H2) for static growth. Biofilms from each of the three donors grew for one, two, three, six or nine days before dispersal. Twenty-three discs per plaque specimen were used to ensure adequate bacterial growth for detection and analysis. Ten discs were collected at day one, six at day two, four at day three, two at day six, and one at day nine. Every 48 hours, growth media was gently removed from wells and replaced by slowly adding a fresh 2 mL of reduced TSB-h. Every donor/substrate/growth-period combination was duplicated.
DNA extraction and 16S rRNA gene sequencing
At the conclusion of the growth period (one, two, three, six or nine days), each disc was removed from the dish and placed into a tube containing 1 mL reduced Ringer’s solution. The tubes were sonicated in a water bath for 30 seconds and vortexed briefly. The remaining biofilm suspension from each experimental condition was spun at 10,000 rpm for 2 minutes to pellet the cells. The supernatant was removed, and the pellet was stored at -20º C for DNA extraction. DNA was extracted from the pellet of each biofilm source for all material/growth-period combinations using microbial DNA extraction kits (Qiagen, Germantown, Maryland, USA) and sent to the Walter Reed Army Institute of Research (WRAIR) for 16S rRNA gene sequencing. MiSeq Reagent Kits (Illumina, San Diego, California, USA) were used for gene sequencing.
Outcome assessment
The alpha and beta diversity values for biofilms grown on each material type were assessed using methods established by Whittaker in 1972 [29,30]. Alpha diversity refers to the species richness within a particular location or on a particular surface. The Shannon Diversity Index (SDI), a parameter incorporating both the number of species in a sample and the abundance of each species, provides an estimate of the alpha diversity. SDI can range in value from unity (indicating one dominant species) to the total number of species in the sample. Species diversity between groups is known as beta diversity. Theoretically, identical groups would exhibit no dissimilarity, producing a beta diversity value of zero. Completely dissimilar groups would register a maximal beta diversity value of one.
Level of analysis
Analyses in this investigation were performed at the operational taxonomic unit (OTU) level. Thus, closely related species were classified together within taxonomic units. This approach contrasts with an amplicon sequence variant (ASV)-level analysis, in which unique sequences are classified as individual species. The approach applied in this study decreased phylogenetic precision but reduced “noise” in the data and permitted assessment of bacterial complexity across experimental groups. To ensure high data fidelity, sequences were quality-filtered and clustered into 97% similarity OTUs, identified to the genus level.
Statistical analyses
Microbiome Insights (Vancouver, British Columbia, Canada) completed the statistical analyses. Sequencing quality was first assessed using FastQC 0.11.5. MiSeq-generated Fastq files were quality-filtered, and bacteria were clustered into 97% similarity operational OTUs using the mothur software package [31]. The SDI was calculated for each experimental group, and a linear mixed-model tested for statistically significant differences. Principal component analysis (PCA) was used to evaluate beta diversity. All microbial profiles were intercompared in a pairwise fashion to establish dissimilarity scores, which were recorded in a distance dissimilarity matrix. Low dissimilarity scores identified similar samples. Abundance-weighted sample pairwise differences were calculated using Bray-Curtis dissimilarity (the ratio of the summed absolute differences in counts to the sum of abundances in the two samples) [32]. Permutational analysis of variance (adonis R function, or Permanova) was used to assess for significant differences in beta diversity associated with substrate and growth period.
Results
Sequence quality determination
The dataset consisted of 56,680 OTUs, excluding those exhibiting fewer than three counts. The samples (all donor/substrate/growth-period combinations) generated an average of 265,002 quality-filtered reads. One sample was eliminated from the analysis due to a low read count. Overall sample quality was favorable for comparison of the experimental groups.
Sequence quality was determined by evaluating the number of quality reads per sample, the quality score across all bases, and the distribution of average quality score across all bases. Quality scores incorporated the base composition and nucleotide distribution, the guanine-cytosine (GC) content distribution, and the duplication rate. An evaluation of the resulting sequence quality data revealed a near-linear correlation between the number of quality reads and the number of days of biofilm growth for the TI- and YSZ-based samples with the most pronounced reduction in quality reads occurring between days six and nine. However, this correlation was not found when biofilms were grown on HA (Figure 1).
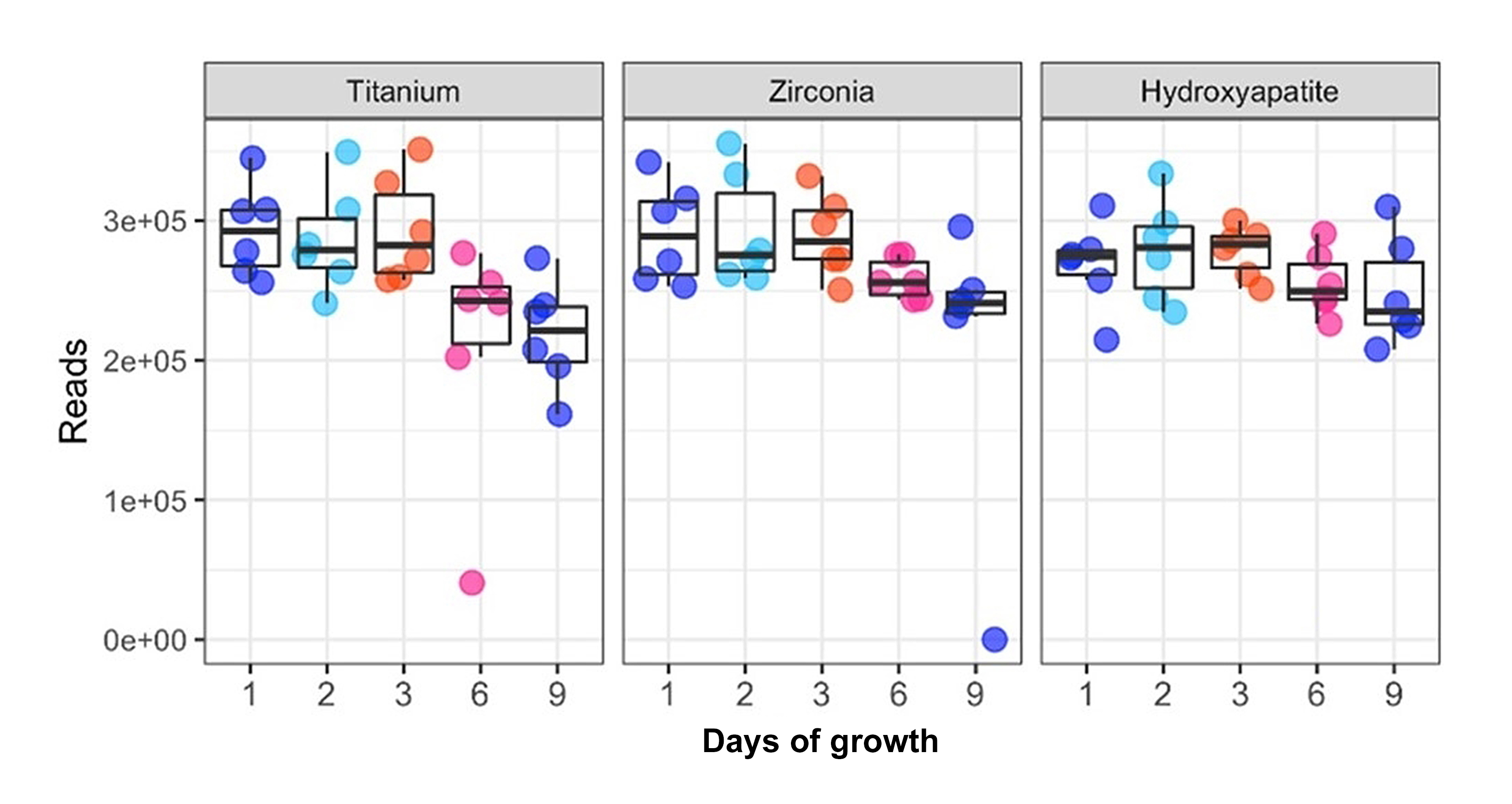
Figure 1: Sequencing quality assessment. Number of quality reads of the 16Sv3v4 region in each sample after discarding poor quality reads. This evaluation confirms adequate sequencing quality for diversity analysis. The number of quality reads per sample was similar among the materials evaluated with a downward trend over nine days of growth. One sample was discarded due to insufficient read data (fewer than 1000 quality reads).
Genome diversity determination
A total of 16 taxa were identified. A qualitative review of the data indicated that the predominant taxa were members of the genera Streptococcus and Veillonella. In terms of abundance these were followed by members of the genera Fusobacterium and Lactobacillus. The least abundant genera included Stomatobaculum and Prevotella. Relative abundances of the predominant phyla and genera were not significantly different across experimental groups, indicating minimal influence of substrate on taxonomic composition. When the SDI was plotted against days of growth, diversity increased over time (Figures 2 and 3). However, biofilms grown on HA surfaces demonstrated significantly higher alpha diversity compared with those formed on TI or YSZ (p<0.0001). Although biofilm complexity tended to increase over time at the genera level, cultures on HA surfaces displayed the greatest diversity at day one, with diversity of biofilms cultured on the three substrates converging at later time points (Figure 2). Notably, members of the genus Streptococcus were abundant at day two on all three materials tested, while members of the genus Fusobacterium were abundant on the TI and YSZ surfaces at day one. Furthermore, each plaque donor exhibited a unique microbiome profile at each time point. Each donor displayed a distinct taxonomic profile dominated by the Streptococcus, Fusobacterium, and Veillonella genera (Figure 4). Dissimilarity among the three groups of samples (HA, TI, and YSZ) was estimated by calculating the beta diversity. When the diversity of all samples in this study were plotted using PCA, the TI and YSZ samples clustered together, and HA samples formed a distinct cluster (Figure 5). Statistically significant differences in beta diversity associated with substrate (p=0.012) and growth period (p=0.001) were noted.
In the PCA, differences in biofilm diversity by substrate tended to vanish over longer growth periods.
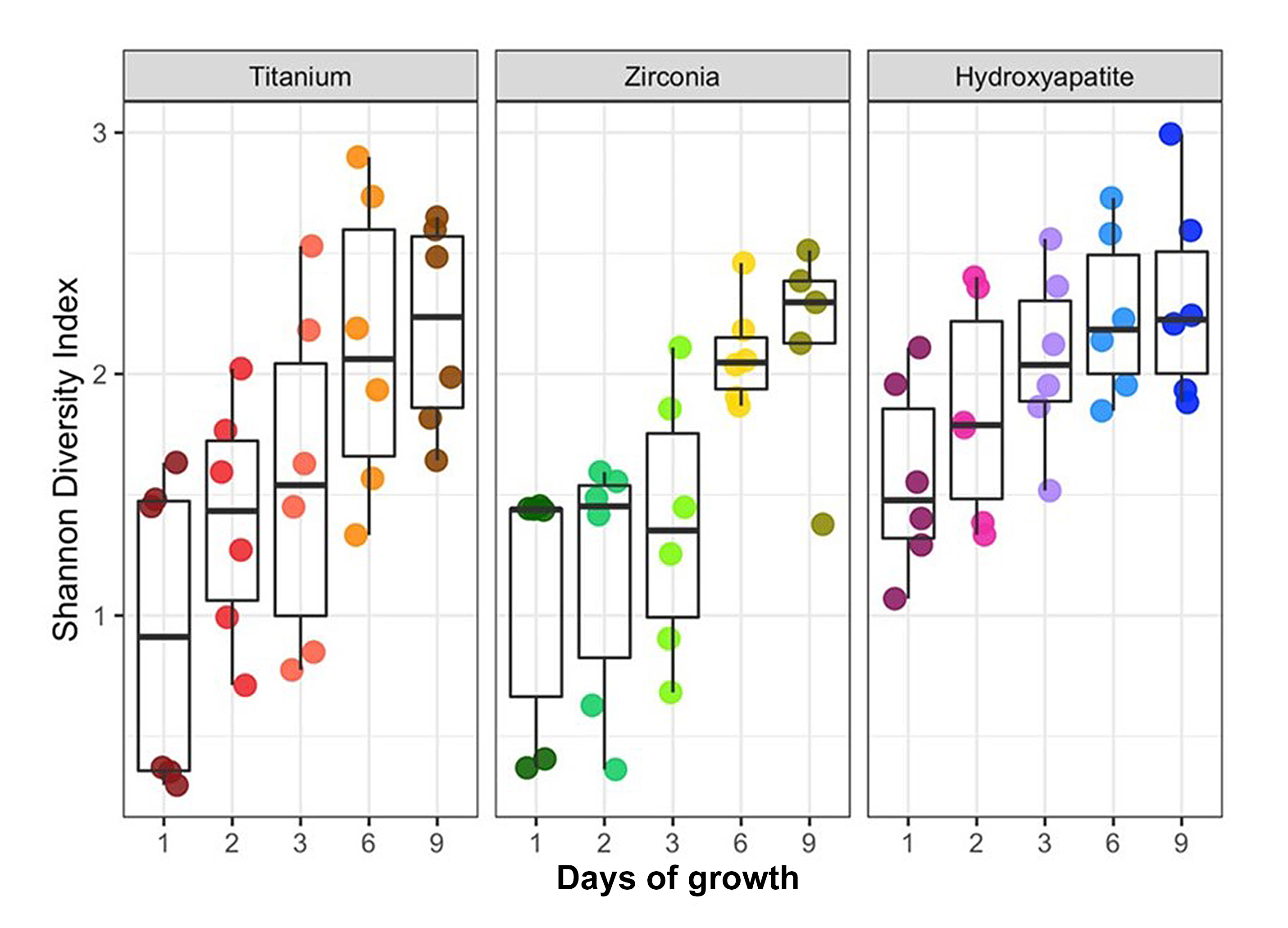
Figure 2: Alpha diversity analysis. Box and whisker plots of the Shannon Diversity Index (SDI) values. Cultures grown on hydroxyapatite exhibited significantly greater alpha diversity than those formed on zirconia or titanium (p<0.0001).

Figure 3: Genera-level microbiome composition by substrate material. For cultures on titanium and zirconia substrates, biofilm complexity increased over time, whereas cultures on hydroxyapatite surfaces were most diverse at day one.

Figure 4: Genera-level microbiome composition by dental plaque donor. Biofilm complexity tended to increase over time. Each specimen exhibited a unique microbial profile. For example, donor 1 exhibited a larger proportion of the genus Fusobacterium compared with donors 2 and 3.
Discussion
In this study, the diversity of bacteria derived from human dental plaque specimens grown on HA, TI, and YSZ was evaluated in vitro, and the potential contribution of each substrate material to species diversity was explored. The results were surprising in that the genomic analysis demonstrated minimal influence of the substrate material on the taxonomic composition of the biofilms (low beta diversity). In other words, at each timepoint, a similar level of diversity was found for each material. These observations suggest that none of the evaluated materials, which are chemically distinct, secrete ions or other substances capable of inhibiting or promoting bacterial growth. However, this hypothesis is countered by the finding of statistically significant differences in SDI values (alpha diversity) occurring over time with the HA discs supporting the most diverse biofilms. This interesting result may indicate that natural teeth support higher microbial diversity levels than the inorganic materials artificially introduced into the oral cavity during dental implant procedures.
In a previous study, investigators evaluated the dynamics of in vitro biofilm formation/maturation on HA, TI, and zirconia discs using three assessment methods—confocal laser scanning microscopy with vital fluorescence, scanning electron microscopy, and quantitative polymerase chain reactions [33]. The discs were inoculated with a combination of six bacterial species selected for the analysis, representing initial, early, intermediate, and late colonizers [33]. The authors found that bacterial adherence and biofilm maturation exhibited similar dynamics, irrespective of the substrate material [33]. This finding is consistent with the bacterial diversity results reported here. Notably, substrate material did appear to influence deposition of extracellular polysaccharide matrix, biofilm thickness, and spatial biofilm organization [33]. These factors may explain the observation of higher alpha diversity among the biofilms grown on HA in the present investigation.

Figure 5: Beta diversity analysis. All profiles were compared in pairwise fashion to determine dissimilarity scores, which were then plotted in a distance dissimilarity matrix. Distance functions produce low dissimilarity scores when comparing similar samples. Abundance-weighted sample pairwise differences were calculated using Bray-Curtis dissimilarities (ratios of the summed absolute differences in counts to the sum of abundances in paired samples). Permutational analysis of variance (adonis R function, or Permanova) determined significant differences in beta diversity associated with substrate (p=0.012) and growth period (p=0.001). Dissimilarity scores for cultures grown on titanium and zirconia clustered together, whereas dissimilarity scores for cultures grown on hydroxyapatite formed a distinct grouping.
At mucosal surfaces throughout the human body, health is often, but not universally, associated with high microbial diversity. While complex microbiota have been found in both inflamed and noninflamed maxillary sinuses, cultures from chronic sinusitis patients often exhibit reduced diversity [34]. Additionally, it is generally accepted that high gut microbial diversity is associated with community stability and health [35,36]. Likewise, some conditions which manifest in severe periodontal destruction have been associated with less diverse microbiomes. This was demonstrated by Moutsopoulos and colleagues who compared the composition of subgingival plaque specimens from patients diagnosed with Leucocyte Adhesion Deficiency I (LAD-I), Localized Aggressive Periodontitis (LAP), and periodontal health using comprehensive 16S rRNA gene-based microarrays [37]. The authors found that health-associated microbial communities exhibited increased diversity, with more numerous, less unique, and less invasive species [37]. In contrast, microbial communities associated with bacterial vaginosis appear significantly more complex than those from healthy controls [38]. Periodontitis results from an exaggerated/unbalanced inflammatory response to bacteria commonly found in the oral cavity, with considerable redundancy in disease-associated species [11,12]. Although specific microbes have been associated with sites of clinical periodontal destruction, accumulation of key microbial gene combinations within the biofilm—rather than presence or absence of particular species—may represent a principal concern in the etiopathogenesis of the disease [11,12].
Conclusions
Under the conditions described, biofilms grown on TI or YSZ appeared significantly less complex than those formed on HA. Limited evidence suggests that high biofilm diversity may support periodontal/peri-implant health. Thus, compared with periodontal health, peri-implant health may be more difficult to establish and maintain, due in part to material characteristics of the transmucosal implant abutment. Observations in the present study underscore the requirement for close monitoring and supportive periodontal care for dental implant patients. In addition, among multiple important considerations in dental implant abutment design, biofilm diversity performance may represent a clinically relevant benchmark. Although additional research is needed, striving to match or exceed the biofilm diversity observed on HA surfaces may positively influence the incidence of peri-implant disease and the effectiveness of treatment.
Author contributions: All authors have contributed substantially to conceptualization of this investigation, drafting the article, critical review, and editing. All authors have approved the final version of the manuscript.
Conflicts of interest: The authors report no financial, economic, or professional interests that may have influenced the design, execution, or presentation of this work.
Disclaimers: The views expressed in this manuscript are those of the authors and do not necessarily reflect the official policy of the United States Government, the Department of Defense, the Defense Health Agency, or Uniformed Services University of the Health Sciences.
Funding: The Defense Health Agency funded this research entirely. The authors received no extramural funding.
References
- Schroeder HE, Listgarten MA. (1997) The gingival tissues: the architecture of periodontal protection. Periodontol 2000. 13: 91-120. [PubMed.]
- Magnusson I, Runstad L, Nyman S, Lindhe J. (1983) A long junctional epithelium‐A locus minoris resistentiae in plaque infection? J Clin Periodontol. 10(3): 333-340. [PubMed.]
- Bartold PM, Walsh LJ, Narayanan AS. (2000) Molecular and cell biology of the gingiva. Periodontol 2000. 24: 28-55. [PubMed.]
- Listgarten MA, Lang NP, Schroeder HE, Schroeder A. (1991) Periodontal tissues and their counterparts around endosseous implants. Clin Oral Implants Res. 2(3): 1-19. [PubMed.]
- Schroeder HE, Listgarten MA. (2003) The junctional epithelium: from strength to defense. J Dent Res. 82(3): 158-161. [PubMed.]
- Sanz M, Herrera D, Kebschull M, Chapple I, Jepsen S, et al. (2020) Treatment of stage I–III periodontitis-The EFP S3 level clinical practice guideline. J Clin Periodontol. 47(Suppl 22): 4-60. [PubMed.]
- Mombelli A. (2003) Periodontitis as an infectious disease: specific features and their implications. Oral Dis. 9(Suppl 1): 6-10. [PubMed.]
- Dye BA. (2012) Global periodontal disease epidemiology. Periodontol 2000. 58: 10-25. [PubMed.]
- Derks J, Tomasi C. (2015) Peri‐implant health and disease. A systematic review of current epidemiology. J Clin Periodontol. 42(Suppl 16): S158-S171. [PubMed.]
- Salvi GE, Cosgarea R, Sculean A. (2017) Prevalence and mechanisms of peri-implant diseases. J Dent Res. 96(1): 31-37. [PubMed.]
- Scannapieco FA, Dongari‐Bagtzoglou A. (2021) Dysbiosis revisited: Understanding the role of the oral microbiome in the pathogenesis of gingivitis and periodontitis: A critical assessment. J Periodontol. 92(8): 1071-1078. [PubMed.]
- Kumar PS. (2021) Microbial dysbiosis: The root cause of periodontal disease. J Periodontol. 92(8): 1079-1087. [PubMed.]
- Vacca-Smith AM, Bowen WH. (1998) Binding properties of streptococcal glucosyltransferases for hydroxyapatite, saliva-coated hydroxyapatite, and bacterial surfaces. Arch Oral Biol. 43(2): 103-110. [PubMed.]
- Listgarten MA. (1994) The structure of dental plaque. Periodontol 2000. 5(1): 52-65. [PubMed.]
- Kolenbrander PE, Palmer Jr RJ, Rickard AH, Jakubovics NS, Chalmers NI, et al. (2006) Bacterial interactions and successions during plaque development. Periodontol 2000. 42(1): 47-79. [PubMed.]
- Haffajee AD, Socransky SS, Patel MR, Song X. (2008) Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 23(3): 196-205. [PubMed.]
- Hirschfeld L, Wasserman B. (1978) A long‐term survey of tooth loss in 600 treated periodontal patients. J Periodontol. 49(5): 225-237. [PubMed.]
- Kaldahl WB, Kalkwarf KL, Patil KD, Molvar MP, Dyer JK. (1996) Long‐term evaluation of periodontal therapy: I. Response to 4 therapeutic modalities. J Periodontol. 67(2): 93-102. [PubMed.]
- Ramfjord SP, Caffesse RG, Morrison EC, et al. (1987) 4 modalities of periodontal treatment compared over 5 years. J Clin Periodontol. 14(8): 445-452. [PubMed.]
- Löe H, Theilade E, Jensen SB. (1965) Experimental gingivitis in man. J Periodontol. 36: 177-187. [PubMed.]
- Araujo MG, Lindhe J. (2018) Peri‐implant health. J Periodontol. 89(Suppl 1): S249-S256. [PubMed.]
- Renvert S, Persson GR, Pirih FQ, Camargo PM. (2018) Peri‐implant health, peri‐implant mucositis, and peri‐implantitis: Case definitions and diagnostic considerations. J Periodontol. 89(Suppl 1): S304-S312. [PubMed.]
- Heitz‐Mayfield LJ, Salvi GE. (2018) Peri‐implant mucositis. J Periodontol. 89(Suppl 1): S257-S266. [PubMed.]
- Schwarz F, Derks J, Monje A, Wang HL. (2018) Peri-implantitis. J Periodontol. 89(Suppl 1): S267-S290. [Ref.]
- Stones DH, Krachler AM. (2016) Against the tide: the role of bacterial adhesion in host colonization. Biochem Soc Trans. 44(6): 1571-1580. [Ref.]
- Berne C, Ducret A, Hardy GG, Brun YV. (2015) Adhesins involved in attachment to abiotic surfaces by Gram‐negative bacteria. Microbiol Spectr. 3(4). [PubMed.]
- Hymes JP, Klaenhammer TR. (2016) Stuck in the middle: fibronectin-binding proteins in Gram-positive bacteria. Front Microbiol. 7: 1504. [Ref.]
- Di Martino P. (2018) Bacterial adherence: much more than a bond. AIMS Microbiol. 4(3): 563-566. [Ref.]
- Whittaker RH. (1972) Evolution and measurement of species diversity. Taxon. 21(2-3): 213-251. [Ref.]
- Gauch Jr HG, Whittaker RH. (1972) Comparison of ordination techniques. Ecology. 53(5): 868-875. [Ref.]
- Mothur. (2024) [Ref.]
- Bray JR, Curtis JT. (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 27(4): 326-349. [Ref.]
- Sánchez MC, Llama-Palacios A, Fernández E, Figuero E, Marín MJ, et al. (2014) An in vitro biofilm model associated to dental implants: structural and quantitative analysis of in vitro biofilm formation on different dental implant surfaces. Dent Mater. 30(10): 1161-1171. [PubMed.]
- Lee JT, Frank DN, Ramakrishnan V. (2016) Microbiome of the paranasal sinuses: update and literature review. Am J Rhinol Allergy. 30(1): 3-16. [PubMed.]
- Bamola VD, Ghosh A, Kapardar RK, Lal B, et al. (2017) Gut microbial diversity in health and disease: experience of healthy Indian subjects, and colon carcinoma and inflammatory bowel disease patients. Microb Ecol Health Dis. 28(1): 1322447. [Ref.]
- Wei H, Dong L, Wang T, Zhang M, Hua W, et al. (2010) Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol Ecol. 73(3): 577-586. [PubMed.]
- Moutsopoulos NM, Chalmers NI, Barb JJ, Abusleme L, Greenwell-Wild T, et al. (2015) Subgingival microbial communities in Leukocyte Adhesion Deficiency and their relationship with local immunopathology. PLoS Pathog. 11(3): e1004698. [Ref.]
- Ling Z, Kong J, Liu F, Zhu H, Chen X, et al. (2010) Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 11: 488. [Ref.]