>Corresponding Author : Michel Leclerc
>Article Type : Research Article
>Volume : 2 | Issue : 1
>Received Date : 25 January, 2022
>Accepted Date : 5 February, 2022
>Published Date : 8 February, 2022
>DOI : https://doi.org/10.54289/JVVD2200105
>Citation : : Leclerc M. (2022) Hla-E Gene from Ophiocomina Nigra (Echinodermata-Invertebrates). Bioinformatics Data. J Virol Viral Dis 2(1): doi https://doi.org/10.54289/JVVD2200105
>Copyright : © 2022 Leclerc M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Research Article | Open Access
Immunology of Invertebrates, 556 Rue Isabelle Romée, 45640 Sandillon France
*Corresponding author: Michel Leclerc, Immunology of Invertebrates, 556 Rue Isabelle Romée, 45640 Sandillon France
Abstract
In the present report, Asterias rubens Fc DNA Sequence was analysed from its transcriptome in bioinformatics; Identities occurred with other sea stars such as Patiria miniata and specially with mammals’ proteins. Identities with Fc receptor mammal IGE was found.
Introduction:
The aim of this work is to analyse Fc DNA sequence which was discovered in 2016 [1] from
Starting material (dna) sequence in 5’-3’:
TCCATTAGGGCAATGAGTGGGACTGCGCGGCTTGGCACAGATCATCCCTTTTCTATCACGACACCTCGAGTCTTTCCACTTGCCGTTGCTAATCTGTAATGCCACACAGTTATTCTCCAATGATTCGACTCCAGACAGCTCAGTTTGCTCTTCTTCGATGAAGTTCGTGTAGTTGACGGGGGAATCGTTTGACCATTTCCAATCGCTTTCGTTGTGTGTATCATGGAGCCCGATCCACACGTCCCTGTCAATTAGGTCGGTAAGAAAATCATTAATTTCTTGGTCAGTGATGGCGACCAGCCTAGCGCCGTCGTATTTAGTGCACTTCTGTTCAGCATCGACCCAGCGTGCTACATCGTCTGGAATCCAGAAGCATTCATCACGGAAGAGATGGCCGTTGTTTAGGCAGTACTGTGGTTGACCACGTACTGTTTGAAGAAGATGAGCTGACCCAATAACCATCATCATCACGAATGGAATCATTGTGAATTTGTTTGAGATACGTCCGATACGTCCGTCCGTAGATGAAAAAACTGCCGAAGTCTCTCACATAATTCCACCAGGCATTGTTGATGCCTTGCTGCTCTATGGTTGATGCTTGGTGGCAGTCCACGAAAGAATGTGCAGTTAGGGAAAGTCCAGCTTGTATATCTC
Bioinformatics data were performed according Marchler-Bauer et al [2-4].
Results:
1. Blastn original sequence: Data base: Standard data bases (nr mainly.)
Optimization: we used highly similar sequences (mega blast)
We recall that molecule type was dna; the query length of 6542 sequences were selected as shown in the table1: significant aligments were found.
Table 1: Significant aligments from A. rubens
| Description | Scientific name | Max score | Total score | Query cover | E. Value | Per. Ident | Acc Len | Accession |
|---|---|---|---|---|---|---|---|---|
| Predicted: Asterias rubens macrophage mannose receptor 1 like (LOC117293835) mRNA | Asterias rubens | 1197 | 1197 | 100% | 0.0 | 99.69% | 1735 | XM-033776291.1 |
| Asterias rubens genome assembly, chromosome 8 | Asterias rubens | 418 | 1162 | 96% | 4e-112 | 10.00% | 21693562 | LR699099.1 |
The corresponding graphic summary is the following:

2. BlastX original sequence:
The query length is 654in this DNA molecule: non-redundant protein sequences were used as Database (nr).
Sequences producing significant aligments: more than 100 sequences were resumed in Table 2.
Table 2: Aligment comparisons between Asterias rubens and Patiria miniata (Asterids-Echinodermata)
| Description | Scientific name | Max score | Total score | Query cover | E. Value | Per. Ident | Acc Len | Accession |
|---|---|---|---|---|---|---|---|---|
| Macrophage mannose receptor 1-like [Asteria rubens] | Asterias rubens | 342 | 342 | 73% | 1e-112 | 99.38% | 510 | XP_033632182.1 |
| uncharacterized protein LOC119734023 isoform X3 [Patiria miniata] | Patiria miniata | 130 | 203 | 75% | 3e-31 | 39.18% | 529 | XP_038063329.1 |
| Macrophage mannose receptor 1-like isoform X2 [Patiria miniata] | Patiria miniata | 129 | 201 | 65% | 1e-30 | 46.32% | 537 | XP_038063328.1 |
| Macrophage mannose receptor 1-like isoform X1 [Patiria miniata] | Patiria miniata | 129 | 201 | 65% | 2e-30 | 43.48% | 547 | XP_038063326.1 |
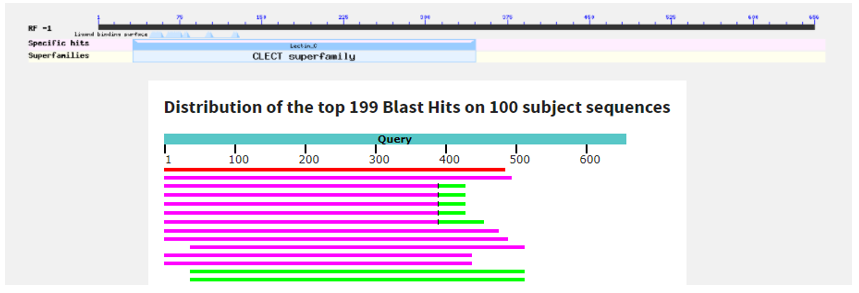
A graphic summary is following as seen below:
3) Putative conserved domains have been detected as shown below:
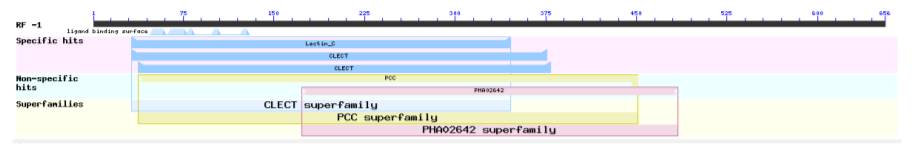
Table 3: Fc gene identities between sea star and mammals
| Name | Accession | Description | Interval | E-value |
|---|---|---|---|---|
| Lectin-C | Pfam00059 | Lectin C-type domain: This family includes both ling and short form of C-type. | 31-354 | 2.33e-19 |
| CLECT | Cd00037 | CLECT: C-type lectin (CTL)/C-type lectin-like (CTLD) domain; protein domains homologous to the carbohydrate-recognition domains (CRDs) of the C-type lectins. This group is chiefly comprised of eukaryotic CTLDs, but contains some, as yet functionally uncharacterized, bacterial CTLDs. Many CTLDs are calcium-dependent carbohydrate binding modules; other CTLDs bind protein ligands, lipids, and inorganic surfaces, including CaCO3 and ice. Animal C-type lectins are involved in such functions as extracellular matrix organization, endocytosis, complement activation, pathogen recognition, and cell-cell interactions. For example: mannose-binding lectin and lung surfactant proteins A and D bind carbohydrates on surfaces (e.g, pathogens, allergens, necrotic, and apoptotic cells) and mediate functions associated with killing and phagocytosis; P (platlet)-, E (endothelial)-, and L (leukocyte)- selectins (sels) mediate the initial attachment, tethering, and rolling of lymphocytes on inflamed vascular walls enabling subsequent lymphocyte adhesion and transmigration. CTLDs may bind a variety of carbohydrate ligands including mannose, N-acetylglucosamine, galactose, N-acetylgalactosamine, and fucose. Several CTLDs bind to protein ligands, and only some of these binding interactions are Ca2+-dependent, including the CTLDs of Coagulation Factors IX/X (IX/X) and Von Willebrand Factor (VWF) binding proteins, and natural killer cell receptors. C-type lectins, such as lithostathine, and some type II antifreeze glycoproteins function in a Ca2+-independent manner to bind inorganic surfaces. Many proteins in this group contain a single CTLD; these CTLDs associate with each other through several different surfaces to form dimers, trimers, or tetramers, from which ligand-binding sites project in different orientations. Various vertebrate type 1 transmembrane proteins including macrophage mannose receptor, endo180, phospholipase A2 receptor, and dendritic and epithelial cell receptor (DEC205) have extracellular domains containing 8 or more CTLDs; these CTLDs remain in the parent model. In some members (IX/X and VWF binding proteins), a loop extends to the adjoining domain to form a loop-swapped dimer. A similar conformation is seen in the macrophage mannose receptor CRD4's putative non-sugar bound form of the domain in the acid environment of the endosome. Lineage specific expansions of CTLDs have occurred in several animal lineages including Drosophila melanogaster and Caenorhabditis elegans; these CTLDs also remain in the parent model. | 31-375 | 946e-18 |
| CLECT | Smart00034 | C-type lectin (CTL) or carbohydrate-recognition domain (CRD); Many of these domain’s function as calcium-dependent carbohydrate binding modules. | 37-378 | 6.87e-15 |
| PCC | TIGR00864 | polycystin cation channel protein; The Polycystin Cation Channel (PCC) Family (TC 1.A.5) Polycystin is a huge protein of 4303aas. Its repeated leucine rich (LRR) segment is found in many proteins. It contains 16 polycystic kidney disease (PKD) domains, one LDL-receptor class A domain, one C-type lectin family domain, and 16-18 putative TMSs in positions between residues 2200 and 4100. Polycystin-L has been shown to be a cation (Na+, K+ and Ca2+) channel that is activated by Ca2+. Two members of the PCC family (polycystin 1 and 2) are mutated in autosomal dominant polycystic kidney disease, and polycystin-L is deleted in mice with renal and retinal defects. Note: this model is restricted to the amino half. | 37-450 | 5.61e-08 |
| PHA02642 | PHA02642 | type lectin-like protein; Provisional. | 172-483 | 5.00e-06 |
The Table 3 represents the identities between Asterias rubens Fc receptor and Mammal IgE Fc receptor:
>Fc fragment of IgE receptor II [Rhinolophus ferrumequinum].
Sequence ID: KAF6306204.1 Length: 290
Range 1: 169 to 284
Score:73.2 bits (178), Expect:3e-11,
Method: Compositional matrix adjust,
Identities:42/119(35%), Positives:69/119(57%), Gaps:5/119(4%).
Query 387 FRDECFWIPDDVARWVDAEQKCTKYDGARLVAITDQEINDFLTDLIDR-DVWIGLHDTHN 211
F+ +C++ + RW+ A C+K G RLV+I QE DFL I R WIGL D +
Sbjct 169 FQRKCYYFGEGAKRWIQARLACSKLQG-RLVSIHSQEEQDFLAKSIHRRGSWIGLRDLNI 227
Query 210 ESDWKWSNDSPVNYTNFIEEEQTELSGVESLENNCVALQISNGKWKDSRCRDR-KGMIC 37
E D+ W +++P++Y+N+ E + G L +CV + +S+G+W D+ C ++ G +C
Sbjct 228 EGDFVWMDENPLDYSNWRPGEPND-GGERGLGEDCVMM-LSSGQWNDAFCGNQLDGWVC 284.
Conclusion:
We retain mainly identities between sea star Asterias rubens Fc receptor and mammal Fc receptor occurs as shown in table-3. Many similitudes have also been observed between A.rubens Fc gene and lectins as CTL. Analogies with macrophage mannose receptor1-like from sea stars were also found. It is interesting to note that mannose sugar binds lectins. We are not surprised to find both.
The most interesting remains, from our side, the identities with mammal Fc receptor genes in conclusion.
References:
- Leclerc M, et al. (2016) Evidence of low affinity immunoglobulin epsilon Fc receptor gene in an invertebrate: The sea star Asterias rubens. Clin. Res. Trials. 2(2): 152-153. [Ref.]
- Marchler-Bauer A, et al. (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45(D1): 200-203. [PubMed.]
- Marchler-Bauer A, et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43(D): 222-226. [PubMed.]
- Marchler-Bauer A, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39(D): 225-229. [PubMed.]
